Momilactone, an allelopathic substance of land plants produced by convergent evolution ~Identification of the first biosynthetic gene cluster in moss~
- Authors
- Lingfeng Mao(Institute of Crop Science, Zhejiang University, PhD student: At the time of research)
Hiroshi Kawaide(Graduate school of Agriculture, Tokyo University of Agriculture and Technology, Professor)
Toshiya Higuchi(Biotechnology Research Center, The University of Tokyo, Master course student:At the time of research)
Meihong Chen(Institute of Crop Science, Zhejiang University, Master course student:At the time of research)
Koji Miyamoto(Department of Biosciences, Teikyo University, Lecturer)
Yoshiki Hirata(Graduate school of Agriculture, Tokyo University of Agriculture and Technology, Master course student:At the time of research)
Honoka Kimura(Graduate school of Agriculture, Tokyo University of Agriculture and Technology, Master course student:At the time of research)
Sho Miyazaki(Graduate school of Agriculture, Tokyo University of Agriculture and Technology, Postdoc:At the time of research)
Miyu Teruya(Biotechnology Research Center, The University of Tokyo, Master course student:At the time of research)
Kaoru Fujiwara(Biotechnology Research Center, The University of Tokyo, Master course student:At the time of research)
Keisuke Tomita(Biotechnology Research Center, The University of Tokyo, PhD student)
Hisakazu Yamane(Department of Biosciences, Teikyo University, Professor:At the time of research)
Ken-ichiro Hayashi(Department of Biochemistry, Okayama University of Science, Professor)
Hideaki Nojiri(Biotechnology Research Center, The University of Tokyo, Professor)
Lei Jia(Institute of Crop Science, Zhejiang University, PhD student)
Jie Qiu(Institute of Crop Science, Zhejiang University, Assistant professor)
Chuyu Ye(Institute of Crop Science, Zhejiang University, Associate Professor)
Michael P. Timko(Department of Biology, University of Virginia, Professor)
Longjiang Fan(Institute of Crop Science, Zhejiang University, Professor)
Kazunori Okada(Biotechnology Research Center, The University of Tokyo, Associate professor)
Significant points
- The biosynthetic genes of the plant defense substance momilactone1) were identified to form a gene cluster on the genome of the lower land plant moss Calohypnum plumiforme.
- Since biosynthetic gene cluster for momilactone is known to exist in the genomes of higher plants such as rice and barnyard grass, this is a first discovery of the biosynthetic gene cluster from lower plants including mosses.
- It is expected that the use of the momilactone biosynthetic gene cluster of moss to create crops resistant to diseases and stress will potentialy lead to new developments in sustainable agriculture.
Summary
The bryophyte moss Calohypnum plumiforme2) often seen with bonsai tree produces momilactone as a chemical compound in allelopathy3) that prevents the growth of surrounding plants. This time, the research group of Biotechnology Research Center, the University of Tokyo, and Tokyo University of Agriculture and Technology, and Zhejiang University jointly discovered that the momilactone biosynthetic genes in the moss constitutes a gene cluster on the moss genome. Previously, the research group reported that rice and barnyard grass in paddy weeds had the momilactone biosynthetic gene cluster, but a biosynthesis gene cluster of secondary metabolites including moss momilactone is the first example of identification in lower plants. In these momilactone-producing plants, the types of biosynthetic genes and their arrangement on the genome are different, suggesting that genes are independently clustered during the evolution of land plants and the ability of momilactone production is presumably acquired by convergent evolution4). Furthermore, the moss-derived biosynthetic pathway was reconstituted in the tobacco, Nicotiana benthamiana, and the production of momilactone A was succeeded for the first time in the momilactone-nonproducing plants. These achievements are not only important clues to understand how land plants have gained a competitive edge to survive using the momilactone biosynthesis gene cluster, but also to use momilactone as a potential natural pesticide in crop production. It is expected to be the first step towards an environment-friendly next-generation agriculture.
Presentation content
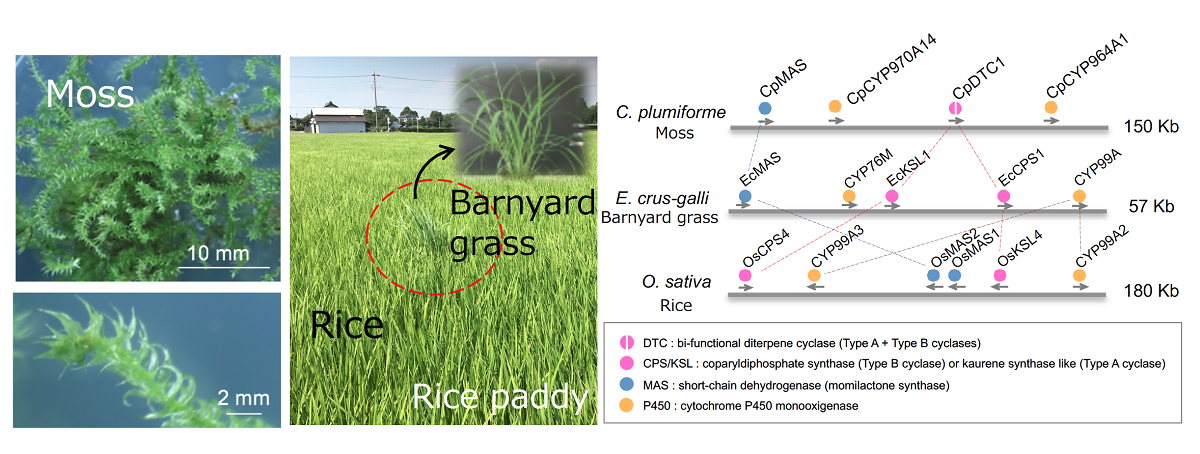
Fig.1 Biosynthetic gene clusters found in momilactone-producing plants
It is known that three evolutionary different plants, lower-plant moss, rice and barnyard grass, produce the momilactones in plant species (left panel). The momilactone biosynthetic gene cluster in the moss consists of four genes, whereas types and arrangement of the corresponding genes in the clusters of rice and barnyard grass are different. P450s belonging to distinctive family are involved in the momilactone synthesis in the moss (right panel).
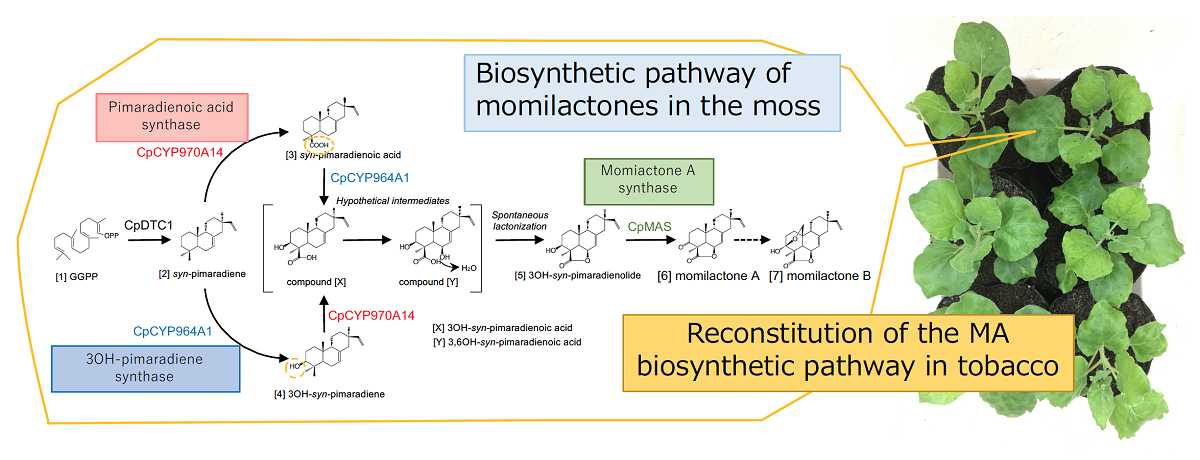
Fig. 2 Reconstitution of the moss momilactone biosynthetic pathway in tobacco.
Momilactone A is heterologously produced by introducing the four momilactone biosynthetic genes(CpDTC1, CpCYP970A14, CpCYP964A1, CpMAS)from the moss into tobacco leaves using agroinfiltration. Investigation of hypothetical intermediates and a step of momilactone B synthesis will be our future works.
Momilactone, a diterpenoid-type phytoalexin5), which is known as an antibacterial compound in rice, was also found in the barnyard grass of paddy weeds, and its biosynthetic genes were known to form clusters on the chromosome (Fig. 1). Momilactone was also reported to be produced as an allelopathic substance in moss, C. plumiforme, a lower land plant bryophytes. As the momilactone biosynthesis gene, CpDTC1 encoding the enzyme responsible for the production of the precursor compound pimaradiene by the two-step cyclization reaction from geranylgeranyl diphosphate (GGPP)6) was the only gene isolated from the moss.
How momilactone is similarly biosynthesized in evolutionarily distant rice and the moss? This is a very interesting mystery when considering emergence of resistance systems in plants utilizing low-molecular chemical substances. To understand the question, it was first necessary to elucidate the whole picture of the momilactone biosynthetic pathway in the moss. In addition, in rice and barnyard grass, where the existence of a momilactone biosynthesis gene cluster has already been shown, the types and arrangements of genes that make up the gene cluster on the chromosome are different, and there seems not to be an evolutionary connection in the formation of the biosynthetic pathway. It was a very important and interesting question whether or not the genes involved in momilactone biosynthesis in the third momilactone-producing plant, C. plumiforme, are clustered on the moss chromosome. Therefore, we started to search for the gene for biosynthesis of momilactone in the moss and to elucidate the biosynthetic pathway.
First, we determined the whole genome sequence of the moss C. plumiforme and comprehensively grasped the genetic information to search for candidates of the momilactone biosynthetic genes. Among the mosses, Physcomitrella patens is often used as a model plant. Our analysis of evolutionary distance comparing the genomic sequences of C. plumiforme and P. patens revealed that these two mosses diverged about 1.7 million years ago, which is almost same with the time when monocotyledons and dicotyledons diverged. This means that, when considering the evolution of moss genomes, the information on C. plumiforme is useful as a new genetic information to be compared, which is different from P. patens. As a result of the gene search with the aid of RNA-seq information obtained in the previous research, CpMAS, which is a homolog of momilactone A synthase gene, and several stress-inducible cytochrome P450 oxidase genes could be picked up in addition to the pimaradiene synthase gene CpDTC1.
In order to investigate whether these genes happened to be adjacent to each other or they function as a momilactone biosynthesis gene, we investigated the enzymatic functions. Regarding the His-CpMAS protein expressed in E. coli and obtained as a purified enzyme, the ability to synthesize momilactone A was revealed in the in vitro reaction using pimaradienolide as a substrate, which is a precursor of momilactone A. The two P450s were named CpCYP970A14 and CpCYP964A1 that retain approximately 90% identity based on comparison with known P450 amino acid sequences, and were analyzed using yeast and tobacco, N. benthamiana. Based on the yeast assays, CpCYP970A14 is a pimaradienoic acid synthase that oxidizes the C19 of pimaradiene to carbonyl, and that CpCYP964A1 also has the activity of hydroxylating the C3 to give 3OH-pimaradiene. Furthermore, when these two P450 genes were simultaneously introduced into tobacco along with the CpDTC1 and CpMAS genes, the GGPP synthase gene required for momilactone synthesis, and the upstream MEP pathway7) genes, we succeeded in reconstructing the synthetic pathway and were able to confirm the production of momilactone A in tobacco (Fig. 2).
From these results, it was proved that the moss momilactone A biosynthetic pathway is composed of four genes and constitutes a gene cluster on the chromosome. This is a first example showing the existence of a biosynthetic gene cluster in lower plants. This result also showed that all the momilactone-producing plants discovered to date have their biosynthetic genes as gene clusters on the chromosome. At the same time, there is no synteny in those momilactone gene clusters, and it is considered that momilactone was accidentally acquired by independent convergent evolution during the evolution of each plant. Alternatively, it may be necessary for genes to be clustered in order to synthesize momilactone. Here, we succeeded in heterologously producing momilactone in tobacco. This is also evidence that the momilactone biosynthesis gene functions without being a cluster. Why the biosynthesis genes of momilactone A are clustered? We would like to keep considering the meaning of clusters by artificially destructuring the cluster or placing genes on different chromosomes. In addition, since momilactone has a useful potential as an antibacterial compound, we challenge to generate new momilactone-producing crops by utilizing the momilactone A biosynthesis gene of the moss. It would be possible to use momilactone as a natural pesticide in next-generation agriculture in the future.
Publishing info
- Journal
- :Proceedings of the National Academy of Sciences of the United States of America
- Title
- :Genomic evidence for convergent evolution of gene clusters for momilactone biosynthesis in land plants
- Authors
- :Lingfeng Mao*, Hiroshi Kawaide*, Toshiya Higuchi*, Meihong Chen, Koji Miyamoto, Yoshiki Hirata, Honoka Kimura, Sho Miyazaki, Miyu Teruya, Kaoru Fujiwara, Keisuke Tomita, Hisakazu Yamane, Ken-ichiro Hayashi, Hideaki Nojiri, Lei Jia, Jie Qiu, Chuyu Ye, Michael P. Timko, Longjiang Fan**, and Kazunori Okada**#
*These authors contributed equally to this work.
** Correspondence
# Lead contact - DOI
- : 10.1073/pnas.1914373117
- URL
- : http://www.pnas.org/cgi/doi/10.1073/pnas.1914373117
Contact information
Lab. of Environmental Biochemistry, Biotechnology Research Center, The University of Tokyo
Kazunori Okada, Ph.D, Associate Professor
Tel:03-5841-3070
Faxl:03-5841-3070
E-mail:ukazokad<atto>mail.ecc.u-tokyo.ac.jp Please replace <atto> @.
Lab.URL:http://park.itc.u-tokyo.ac.jp/biotec-res-ctr/kampo/
Grossary
- 1) Momilactone
An antibacterial compound found in husks of rice, a natural defense substance that has the activity of suppressing the growth of pathogens such as blast fungus. It is considered to give rice competitiveness to other plants because it also has a growth inhibitory effect on plants. Due to its broad range of biological activities, it is expected to be used as a natural pesticide. - 2) The moss Calohypnum plumiforme
It is a moss belonging to the Calohypnum genus in bryophytes. It is widely seen in Asia and is distributed for gardening in Japan and lives everywhere. - 3) Allelopathy
A general term for a biological phenomenon in which an organism produces biochemicals that influence the germination, growth, survival, and reproduction of other organisms. These biochemicals are known as allelochemicals, which have beneficial (positive allelopathy) or detrimental (negative allelopathy) effects on the target organisms and the community. - 4) Convergence evolution
An evolutionary phenomenon in which groups of organisms that have no commonality of genetic inheritance from ancestors individually acquire a common trait (here, the ability to synthesize momilactone) that gives any benefit to each organism through independent natural selection. - 5) Diterpenoid-type phytoalexin
Phytoalexin is a low-molecular compound produced by plants in response to various environmental stresses. There are various types of compounds for each plant species. Phytoalexins in rice are classified into diterpenoid-type momilactones, phytocasanes and flavonoid-type sakuranetin according to their chemical structures and biosynthetic pathways. - 6) Geranylgeranyl diphosphate (GGPP)
GGPP is a C20 prenyl diphosphate harboring an important role as a starting material for diterpenoids. It is essential for the biosynthesis of the plant hormone gibberellins and the photosynthetic pigment chlorophylls other than phytoalexins. GGPP is mainly synthesized in chloroplasts and then undergoes reactions such as cyclization to transform into various diterpene compounds. - 7) MEP pathway
This metabolic pathway functions as a source in the production of plant secondary metabolites and constitutes the upstream pathway of terpenoids. It consists of seven genes responsible for the synthesis of terpenoids through methylerythritol phosphate (MEP). These enzymes are translocated into the chloroplast, and catalyze the conversion of pyruvate and glyceraldehyde triphosphate, the primary metabolites from glycolysis, to isopentenyl diphosphate and its isomer, dimethylallyl diphosphate, both of which are the basic building blocks of terpenoids.

